
For the gDNA digestion, a restriction enzyme is chosen to generate fragments of suitable lengths that can self-ligate. In a conventional workflow for studying unknown sequences of genomic DNA, restriction digestion and ligation precede inverse PCR, which is then followed by sequencing of the PCR amplicon. Today, inverse PCR is routinely employed in site-directed mutagenesis to replicate a target plasmid while introducing desired mutations. The method is known as inverse PCR because the primers are designed to extend away from each other rather than toward each other as in regular PCR. Inverse PCR is helpful for investigating the promoter sequence of a gene oncogenic chromosomal rearrangements such as gene fusion, translocation, and transposition and viral gene integration. Inverse PCR was originally designed to determine sequences of adjacent unknown regions. Hyperthermostable DNA polymerases are also advantageous for GC-rich PCR, since a higher denaturation temperature (e.g., 98☌ instead of 95☌) may facilitate strand separation and PCR amplification (learn more about PCR cycling). Highly processive DNA polymerases are beneficial for GC-rich PCR, because of their strong binding to the templates during primer extension ( Figure 6B). These reagents often lower the primer T m, so the annealing temperature should be adjusted accordingly. To overcome strong GC interactions, the most common approach relies on PCR additives or co-solvents such as DMSO to help DNA denature ( Figure 6A). To amplify GC-rich targets, the double-stranded template must be separated for the primers to bind and DNA polymerase to read through the sequence. Thus, GC-rich sequences can cause DNA polymerases to “stutter” along templates and interrupt DNA synthesis. GC-rich sequences can also be involved in secondary structures. In this manner, desired PCR products are selectively increased with little or no amplification of nonspecific targets over the course of PCR ( Figure 2).ĭNA templates containing high GC content (>65%) can be difficult to amplify because of the stronger hydrogen bonds between G and C bases.
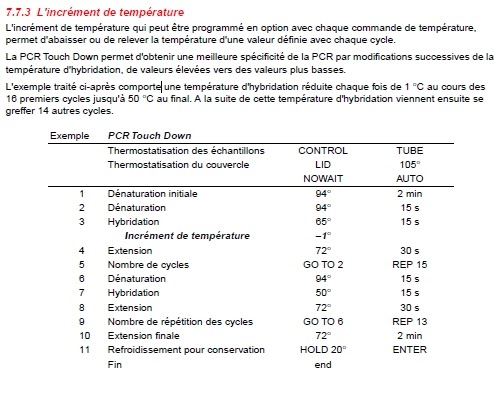
Once the annealing temperature reaches, or “touches down”, at the optimal temperature (usually 3–5☌ lower than the lowest primer T m), it is maintained throughout the remaining cycles for primer annealing. To overcome this challenge, the annealing temperature is often decreased 1☌ at every cycle of the initial few cycles to produce a sufficient yield of the desired amplicon. While preventing primer-dimers and nonspecific primer binding, the higher annealing temperatures may result in lower PCR yield due to increased dissociation of primers from their intended target. As such, higher annealing temperatures reduce nonspecific PCR products and promote specific amplification at the start of PCR (learn more about PCR annealing step). Higher temperatures help destabilize the formation of primer-dimers and nonspecific primer-template complexes, thus minimizing undesirable amplification. In touchdown PCR, the annealing temperature of the first few cycles is set to be a few degrees higher than the highest melting temperature (T m) of the primers. Another approach to promoting specificity is to modify the PCR cycling parameters.


 0 kommentar(er)
0 kommentar(er)
